Task 2 phylogenetic analysis of factor B and C2 sequences
As part of your project, you are required to analyse the complement factor B (FB) and C2 proteins from a number of species. These proteins are part of the immune complement system, a large group of proteins that interact in a cascade manner when activated to initiate an inflammatory response and neutralise pathogens. As part of your introduction, you should discuss the different but analogous roles of factor B and C2 in the complement cascade.
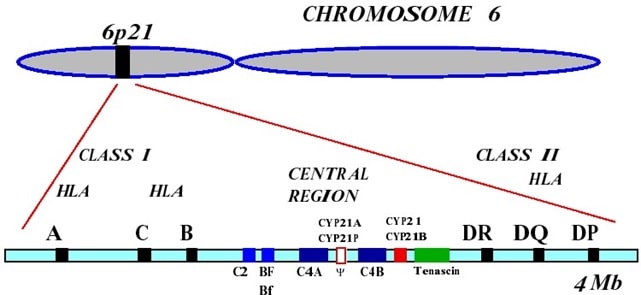
Both the C2 and FB genes in humans are located close to each other on the short arm of chromosome 6 (6p – see figure below). They occur within the central region of the major histocompatibility complex (MHC) along with several other genes that are present in duplicated forms (complement C4 and CYP21). Sequence analysis of both the genes and the proteins they encode show that C2 and FB share similarities which support the hypothesis that these two genes arose by duplication of an ancestral FB like gene early during vertebrate evolution.
Tasks to complete:
- Download representative sequences from each species in which the two genes have been identified and reported -‐ A list of Uniprot sequence headers has been provided to make this task easier.
- Multiply align the sequences using ClustalX or Muscle (You can use SeaView to align using Muscle)
- Generate phylogenetic trees using neighbor-‐joining (NJ) and maximum likelihood (ML) methods.
- Analyse the trees obtained and report on your findings
Software required:
- ClustalX 2.0x – Download from the Bioinformatics Blackboard site or
- SeaView -‐ Download from the Bioinformatics Blackboard site (provides alignment with Muscle)
- MEGA (http://www.megasoftware.net/)
- PhyML –use SeaView (under Tree menu)
Download complement factor B and C2 sequences
Initially you will need to retrieve factor B and C2 sequences from a public database. Note that the factor B and C2 sequences available in the database will often be referred to as precursor or preproprotein sequences. This just means that the sequence represents the product before post-‐translational modification. It is fine to use these precursor sequences for your alignment.
You are provided with a list of Uniprot fasta headers.
Recommended strategies:
- Go to UniprotKB (uniprot.org) and search the database for ‘complement factor B’. Look for entries that correspond to the headers in the reference file. These should be mostly around 750bp in length, although some of the sequences from fish and pre-‐vertebrates are longer. Save the relevant entries in Fasta Format. Record the Accession and Uniprot ID, gene name, organism and length of each sequence in a table.
- You may not retrieve all available factor B and C2 sequences using a search for ‘Complement factor B’. Repeat the search using ‘Complement C2’ to find remaining entries. Record the Accession and Uniprot ID, gene name, organism and length of each sequence in a table.
- Add the predicted proteins for sheep (Ovis aries) complement FB and C2 from the FGENESH+ refined predictions in the genomic sequence FJ985872
NB. Aliases for complement factor B include:
- Complement factor B precursor
- Complement factor B preproprotein
- B-‐factor, properdin or properdin factor B
- Complement component factor B
- Bf
- Complement component Bf/C2 (fish species and lower)
Do not retrieve Factor B like fragments from species (there are many), make sure the sequences are at least 650 aa (the length will vary, tending to increase in lower organisms because of additional SUSHI domains). You will note that many of the fish sequences (and the newer ones from the horseshoe crab, sea anemone and amphioxus) are referred to as Bf/C2 sequences since it is still not known for certain when the factor B gene duplicated to form the gene orthologous to the mammalian C2 gene. Also, there may be other (independent) duplication events involving these genes in fish and other species lower than mammals. Check the database records – genes may be referred to as Bf-‐1, Bf-‐2 or Bf/C2A, Bf/C2B etc. Please note that although there are numerous isoforms for complement factor B and C2 reported in mammals (including humans), these are due to alternate transcripts, not duplicate genes. Choose the isoform 1 if you see a sequence identified as such (These will be in your reference list). In mammals, choose one representative sequence from each mammalian species for complement factor B and one representative sequence for complement component 2 (C2). The chosen sequence should be ~750-‐760 aas.
Your search for complement C2 should turn up a number of hits to Complement C2 sequences that the first search missed.
All sequences should be placed in a single fasta-‐formatted datafile. You may wish to add an abbreviated name at the beginning of each sequence header just after the ‘>’. This will help when trying to identify sequences in the alignment and in the phylogenetic trees. You can use the international convention, 1st two letters of the genus, 1st two letters of the species, then add an underscore and something that indicates which gene it is.
Example names:
|
Identifier |
Gene |
|
HOSA_Bf |
human factor B |
|
MUMU_Bf |
mouse factor B |
|
HOSA_C2 |
human C2 |
|
CYCA_BC2A2 |
Carp Bf/C2 gene A isoform 2 |
|
CYCA_BC2B |
Carp Bf/C2 gene B |
Include a table in your report listing the sequences downloaded for analysis.
|
Species |
Common name |
Gene |
Uniprot Accession |
Identifier |
Length No. of AAcids |
|
Homo sapiens |
Human |
FB |
P00751 |
HOSA_Bf |
764 |
|
Gorilla gorilla |
Gorilla |
FB |
Q864V9 |
GOGO_Bf |
764 |
|
Pan troglodytes |
Chimp |
C2 |
Q8SQ74 |
PATR_C2 |
752 |
|
Oncorhynchus mykiss |
Rainbow Trout |
Bf/C2 |
F1DRC4 |
ONMY_BfC2 |
749 |
Etc.
You will have to search for the common name for a number of the species. Try google, or search the taxonomy database at NCBI. The common name will give you more of a clue about where the organism sits taxonomically (ie within the chart in Appendix 2), which will probably help you with your analysis of the trees.
Multiple sequence alignment of FB and C2 sequences
If you use Clustalx, I recommend that you setup the multiple alignment parameters as follows: Output format options -‐ Aln in Fasta and format Pairwise and multiple alignment options – default gap penalties, BLOSUM matrices
An alternative is to use SeaView to align the sequences, choosing muscle as the alignment option (the default). It is difficult to vary the default alignment options in SeaView, so don’t try to change them. You can use SeaView to save the alignment in various output formats afterwards.
Examine the alignment. Note areas at the front and end of the alignment where there is no data for some sequences (because of differences in sequence length), as well as areas where there are large or non-‐uniform gaps. You could remove these using a software package like GBLOCKS (http://molevol.cmima.csic.es/castresana/Gblocks_server.html). However, MEGA automatically deletes (ignores) positions with gaps, so it is not necessary to remove them when using this software for your phylogenetic analysis.
Phylogenetic Analysis of FB and C2 alignment
Neighbor Joining
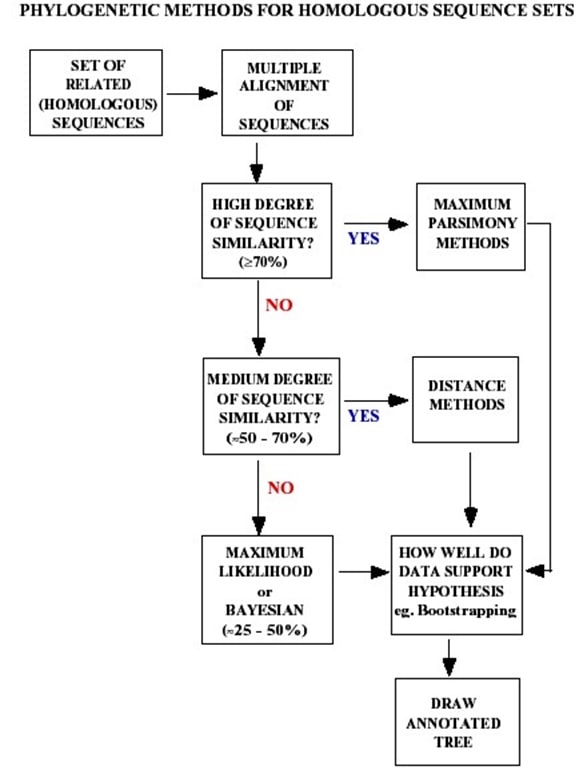
First, use MEGA to generate bootstrapped NJ trees. You will first have to use the Alignment Explorer to export the fasta formatted alignment into the MEGA format. Create a tree using a substitution model of your choice. You may wish to test for the best model before you run the analysis (under Models). Note that when you choose a substitution model (e.g. JTT or WAG), the bootstrap tree will take a while to generate (~30 minutes). Also, generate a proportional distance (p-‐distance) matrix so that you will have some idea of the diversity of the sequences.
You should set the outgroup (root) to the most distantly related species (which is it? If you don’t know, FIND OUT! There is a taxonomic chart to help in Appendix 2. Root the tree on the outgroup, then create an image of each tree for your report (a rectangular cladogram is best, showing bootstrap values on the bootstrapped trees). Save the captions in order to help label your figures. The cited references belong in the references section of your report – reformat both citations and references into the style that you have chosen for your report.
Maximum Likelihood
MEGA5 can be used to generate ML trees (choose the Maximum likelihood option under Phylogeny). Before you do this, use determine the best model to use for your sequence set (Models menu). You should bootstrap the ML tree (if time permits), but choose no more than 100 as the number of bootstraps. Also, be prepared to wait, so do this at home! Reroot the generated tree on the oldest organism. If you have bootstrapped the tree, be sure to check the consensus tree for significant differences to the original tree.
PhyML is available as a tree-‐building method in SeaView (Under the Tree menu) and is faster than the ML option in MEGA5. It provides aLRT support values by default, which are as reliable as bootstrap values and interpreted in a similar way. To use the program, open the CFB and C2 sequence file in SeaView. Align the sequences if they are not already aligned. Choose PhyML under the Tree menu and use the default values to begin with. It will take a while to generate the tree, a progress window will appear, then the tree when the analysis is finished. You will not see the aLRT values on the tree shown, you must first tick the ‘bootstrap’ box. Reroot the tree (click the circle next to this option) by choosing the sequence or sequence clade that you wish to root the tree on, then restore the Full tree before saving. Seaview generated trees can be saved as PDF documents. Try generating trees with and without rate variation. Leave the number of substitution rate categories at 4 and choose to estimate the gamma shape parameter. Be sure to root the tree on the same organism(s) that you have used for previous trees.
Hints for the discussion and critical appraisal
Analysis of the phylogenetic trees
Comment on the information contained in the NJ tree. Your discussion may include the grouping of the FB and C2 sequences, the pattern of the clades (groups) present in the tree (ie the tree topology) and their congruence with other phylogenetic classifications, and identification of some fish sequences as either FB or C2 like. Take note of the bootstrap values and identify branches/nodes in which you have the most and least confidence. Compare the original tree with the consensus bootstrap tree.
ML trees
If you have more than one tree (e.g. from multiple analyses using different parameter settings), pick one ‘best’ tree from each method to compare with each other and the NJ tree. Comment on the tree topology, as you did for the NJ tree, and also comment on differences and similarities between the trees. If you bootstrapped the ML tree in MEGA5, comment on differences between the original and consensus tree.
Presenting trees in your report
Present one tree in your results section that you think is the best tree for determining the probable time of the CBF gene duplication that led to the evolution of the current mammalian C2 gene. Annotate this tree. Other trees should be presented in appendices so that I can refer to them if necessary.
You should have determined the average distance among the sequence set in MEGA5; from this determine the average percent identity, and comment on what is probably the ‘best’ method to use for this sequence set after referring to Appendix 1.
Evolution of FB/C2 genes
Identify any obvious paralogous and orthologous relationships among the complement factor B and C2 sequences on the tree. Does your analysis of the dataset concur with the generally accepted view that complement C2 arose from a duplication of the factor B gene during early vertebrate evolution? Can you suggest when this may have occurred? Are all of the pre-mammalian sequences Factor B-‐like, or do some appear to be more closely related to C2? Is there evidence of independent factor B gene duplication events in some species?
Recommended References for this analysis
Masura, N and Kimura, A. 2006. Genomic view of the evolution of the complement system. Immunogenetics. 58:701-‐713.
Zhu, Y. et al. 2005. The ancient origin of the complement system. EMBO. 24:382-‐394. Fujita, T, Matsushita, M and Endo, Y. 2004. The lectin-‐complement pathway – its role in innate immunity and evolution. Immunological Reviews. 198:185-‐202
Nakao, M. et al. 2002. Diversity of complement factor B/C2 in the common carp: three isotypes of B/C2-‐A expressed in different tissues. Dev and Comp Immunol. 25:533-‐541.
Gongora, R. Figueroa, F. and Klein, J. 1998. Independent Duplications of Bf and C3 Complement Genes in the Zebrafish. Scand. J. Immunol. 48:651–658.
Nakao, M. et al. 1998. Two Diverged Complement Factor B/C2-‐Like cDNA Sequences from a Teleost, the Common Carp (Cyprinus carpio). J Immunol. 161: 4811–4818.
Kato Y, et al. 1995. Duplication of the MHC-‐linked Xenopus complement factor B gene. Immunogenetics. 42(3):196-‐203.
Appendix 1
Appendix 2 – Taxonomy Chart
|
Phylum |
Order |
Class |
Subclass |
Common Name |
Species |
Divergence ~ Mya | |
|
CHORDATA SUBPHYLUM VERTEBRATA |
TETRAPODA |
HOMEO THERMS |
Mammalia |
Mammals Primate |
Man |
Homo Sapiens |
0 |
|
Chimpanzee |
Pan troglodytes |
25 | |||||
|
Orangutan |
Pongo pygmaeus |
30 | |||||
|
Rodent |
Mouse |
Mus musculus |
100 | ||||
|
Aves |
Birds |
Chicken |
Gallus gallus |
300 | |||
|
POIKILO THERMS |
Reptilia |
Reptiles |
Snakes, Skinks Turtles, Alligators |
Naja kaouthia |
300 | ||
|
Amphibia |
Amphibians |
Frogs, Toads |
Xenopus laevis |
380 | |||
|
FISHES (PISCES) |
Osteichthyes (Bony fishes) |
Perciformes |
Tuna |
400 | |||
|
Salmonids |
Rainbow Trout |
Oncorhynchus mykiss |
400 | ||||
|
Medakafish |
Oryzias latipes |
400 | |||||
|
Cypriniformes |
Zebrafish |
Danio rerio |
400 | ||||
|
Carp |
Cyprinus carpio |
400 | |||||
|
Chondrichthyes |
Cartilaginous fishes |
Dogfish |
Triakis scyllium |
470 | |||
|
Nurse Shark |
Ginglymostoma cirratum | ||||||
|
Placoderms |
Early jawless fishes - mostly armoured |
Extinct order | |||||
|
DEUTEROSTOMES |
Agnathans |
Lamprey |
Lethenteron japonicum |
530 | |||
|
Hagfish |
Eptatretus stoutii |
550 | |||||
|
PROTO CHORDATES |
Cephalochordates |
Branchiostomes |
Amphioxus |
Amphioxus | |||
|
Urochordates |
Ascidians |
Tunicate |
Halocynthia roretzi |
650 | |||
|
Hemichordates | |||||||
|
Echinoderms |
Purple sea urchin |
Strongylocentrotus purpuratus |
700 | ||||
|
PROTOSTOMES |
Annelids | ||||||
|
Molluscs | |||||||
|
Nematodes |
Worm |
750 | |||||
|
Arthropods |
Insect |
Fruit fly |
Drosophila melanogaster |
750 | |||


